LatinHypercubeSearch¶
-
class
getml.hyperopt.LatinHypercubeSearch(model, param_space=None, seed=None, session_name='', n_iter=30)¶ Bases:
getml.hyperopt.hyperopt._BaseSearchLatin hypercube sampling of the hyperparameters.
Uses a multidimensional, uniform cumulative distribution function to drawn the random numbers from. For drawing n_iter samples, the distribution will be divided in n_iter`*`n_iter hypercubes of equal size (n_iter per dimension). n_iter of them will be selected in such a way only one per dimension is used and an independent and identically-distributed (iid) random number is drawn within the boundaries of the hypercube.
Like in
RandomSearchthe sampling is based on a purely statistical algorithm and does not incorporate the results of previous evaluations.Examples
population_table_training, peripheral_table = getml.datasets.make_numerical( random_state = 132) population_table_validation, _ = getml.datasets.make_numerical( random_state = 133) population_placeholder = population_table_training.to_placeholder() peripheral_placeholder = peripheral_table.to_placeholder() population_placeholder.join(peripheral_placeholder, join_key = "join_key", time_stamp = "time_stamp" ) feature_selector = getml.predictors.LinearRegression() predictor = getml.predictors.XGBoostRegressor() m = getml.models.MultirelModel( population = population_placeholder, peripheral = peripheral_placeholder, feature_selector = feature_selector, predictor = predictor, name = "multirel" ).send() param_space = { 'num_features': [80, 150], 'regularization': [0.3, 1.0], 'shrinkage': [0.1, 0.9] } l = getml.hyperopt.LatinHypercubeSearch( model = m, param_space = param_space, seed = int(datetime.datetime.now().timestamp()*100), session_name = 'test_search', n_iter = 10 ) l.fit( population_table_training = population_table_training, population_table_validation = population_table_validation, peripheral_tables = peripheral_table ) l.get_scores()
- Parameters
model (Union[
MultirelModel,RelboostModel]) – Base model used to derive all models fitted and scored during the hyperparameter optimization. Be careful in constructing it since only those parameters present in param_space too will be overwritten. It defines the data schema, any hyperparameters that are not optimized, and contains the predictor which will - depending on the parameter space - will be optimized as well.param_space (dict, optional) –
Dictionary containing numerical arrays of length two holding the lower and upper bounds of all parameters which will be altered in model during the hyperparameter optimization. To keep a specific parameter fixed, you have two options. Either ensure it is not present in param_space but in model, or set both the lower and upper bound to the same value. Note that all parameters in the
modelsandpredictorsdo have appropriate default values.If param_space is None, a default space will be chosen depending on the particular model and model.predictor. These default spaces will contain all parameters supported for the corresponding class and are listed below.
-
{ 'grid_factor': [1.0, 16.0], 'max_length': [1, 10], 'min_num_samples': [100, 500], 'num_features': [10, 500], 'regularization': [0.0, 0.01], 'share_aggregations': [0.01, 1.0], 'share_selected_features': [0.1, 1.0], 'shrinkage': [0.01, 0.4] }
-
{ 'max_depth': [1, 10], 'min_num_samples': [100, 500], 'num_features': [10, 500], 'reg_lambda': [0.0, 0.1], 'share_selected_features': [0.1, 1.0], 'shrinkage': [0.01, 0.4], }
LinearRegressionandLogisticRegression{ 'predictor_learning_rate': [0.5, 1.0], 'predictor_lambda': [0.0, 1.0] }
XGBoostClassifierandXGBoostRegressor{ 'predictor_n_estimators': [10, 500], 'predictor_learning_rate': [0.0, 1.0], 'predictor_max_depth': [3, 15], 'predictor_reg_lambda': [0.0, 10.0] }
To distinguish between the parameters belonging to the model from the ones associated with its predictor, the prefix ‘predictor_’ has to be added to the latter ones.
-
seed (Union[int,None], optional) – Seed used for the random number generator that underlies the sampling procedure to make the calculation reproducible. Due to nature of the underlying algorithm this is only the case if the fit is done without multithreading. To reflect this, a seed of None does represent an unreproducible and is only allowed to be set to an actual integer if both
num_threadsandn_jobsinstance variables of thepredictorandfeature_selectorin model - if they are instances of eitherXGBoostRegressororXGBoostClassifier- are set to 1. Internally, a seed of None will be mapped to 5543. Range: [0,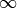 ]
]session_name (string, optional) –
Unique ID which will be both used as prefix for the
nameparameter of all models fitted during the hyperparameter optimization and directly inserted intosession_name. It will be used as a handle to load the constructed class from the getML engine.If session_name is empty, a default one based on the current date and time will be created.
Using a session_name all models trained in the engine during the hyperparameter optimization, which are based on the provided model, can be identified unambiguously.
n_iter (int, optional) – Number of iterations in the hyperparameter optimization and thus the number of parameter combinations to draw and evaluate. Range: [1,
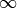 ]
]
- Raises
KeyError – If an unsupported instance variable is encountered (via
validate()).TypeError – If any instance variable is of wrong type (via
validate()).ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical) (via
validate()).ValueError – If not
predictoris present in the provided model.
Methods Summary
fit(population_table_training, …[, score])Launches the hyperparameter optimization.
Get a list of all models fitted during the hyperparameter optimization.
Get a dictionary of the score corresponding to all models fitted during the hyperparamer optimization.
validate()Checks both the types and the values of all instance variables and raises an exception if something is off.
Methods Documentation
-
fit(population_table_training, population_table_validation, peripheral_tables, score=None)¶ Launches the hyperparameter optimization.
The optimization itself will be done by the getML software and this function returns immediately after constructing the request and checking whether population_table_training and population_table_validation do hold the same column names using
_validate_colnames().In every iteration of the hyperparameter optimization a new set of hyperparameters will be drawn from the param_space member of the class, those particular parameters will be overwritten in the base model and it will be renamed, fitted, and scored. How the hyperparameters themselves are drawn depends on the particular class of hyperparameter optimization.
The provided
DataFramepopulation_table_training, population_table_validation and peripheral_tables must be consistent with thePlaceholdersprovided when constructing the base model.- Parameters
population_table_training (
DataFrame) – The population table that models will be trained on.population_table_validation (
DataFrame) – The population table that models will be evaluated on.peripheral_tables (
DataFrame) – The peripheral tables used to provide additional information for the population tables.score (string, optional) –
The score with respect to whom the hyperparameters are going to be optimized.
Possible values for a regression problem are:
Possible values for a classification problem are:
cross_entropy(default)
- Raises
TypeError – If any of population_table_training, population_table_validation or peripheral_tables is not of type
DataFrame.KeyError – If an unsupported instance variable is encountered (via
validate()).TypeError – If any instance variable is of wrong type (via
validate()).ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical) (via
validate()).
-
get_models()¶ Get a list of all models fitted during the hyperparameter optimization.
- Returns
List of all models fitted during the hyperparameter optimization.
- Return type
list
- Raises
Exception – If the engine yet reports back that the operation was not successful.
KeyError – If an unsupported instance variable is encountered (via
validate()).TypeError – If any instance variable is of wrong type (via
validate()).ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical) (via
validate()).
-
get_scores()¶ Get a dictionary of the score corresponding to all models fitted during the hyperparamer optimization.
- Returns
All score fitted during the hyperparameter optimization. Each field adheres the following scheme:
{"model-name": {"accuracy": [list_of_scores], "auc": [list_of_scores], "cross_entropy": [list_of_scores], "mae": [list_of_scores], "rmse": [list_of_scores], "rsquared": [list_of_scores]}
For more information regarding the scores check out
getml.models.scores(listed under ‘Variables’).- Return type
dict
- Raises
Exception – If the engine yet reports back that the operation was not successful.
KeyError – If an unsupported instance variable is encountered (via
validate()).TypeError – If any instance variable is of wrong type (via
validate()).ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical) (via
validate()).
-
validate()¶ Checks both the types and the values of all instance variables and raises an exception if something is off.
Examples
population_table, peripheral_table = getml.datasets.make_numerical() population_placeholder = population_table.to_placeholder() peripheral_placeholder = peripheral_table.to_placeholder() population_placeholder.join(peripheral_placeholder, join_key = "join_key", time_stamp = "time_stamp" ) feature_selector = getml.predictors.LinearRegression() predictor = getml.predictors.XGBoostRegressor() m = getml.models.MultirelModel( population = population_placeholder, peripheral = peripheral_placeholder, feature_selector = feature_selector, predictor = predictor, name = "multirel" ).send() param_space = { 'num_features': [80, 150], 'regularization': [0.3, 1.0], 'shrinkage': [0.1, 0.9] } l = getml.hyperopt.LatinHypercubeSearch( model = m, param_space = param_space, seed = int(datetime.datetime.now().timestamp()*100), session_name = 'test_search' ) l.optimization_burn_ins = 240 l.model.num_threads = 2 l.validate()
- Raises
KeyError – If an unsupported instance variable is encountered.
TypeError – If any instance variable is of wrong type.
ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical).
Note
This method is called at end of the __init__ constructor and every time a method is communicating with the getML engine.
To directly access the validity of single or multiple parameters instead of the whole class, you can used
getml.helpers.validation.validate_hyperopt_parameters().