GaussianHyperparameterSearch¶
-
class
getml.hyperopt.GaussianHyperparameterSearch(model, param_space=None, session_name='', n_iter=100, ratio_iter=0.75, optimization_algorithm='nelderMead', optimization_burn_in_algorithm='latinHypercube', optimization_burn_ins=15, seed=None, surrogate_burn_in_algorithm='latinHypercube', gaussian_kernel='matern52', gaussian_optimization_algorithm='nelderMead', gaussian_optimization_burn_in_algorithm='latinHypercube', gaussian_optimization_burn_ins=50)¶ Bases:
getml.hyperopt.hyperopt._BaseSearchBayesian hyperparameter optimization using a Gaussian process.
In contrast to
LatinHypercubeSearchandRandomSearchthe Bayesian hyperparameter search is not a purely statistical algorithm. After a burn-in period (purely statistically), a Gaussian process is used to pick the most promising parameter combination to be evaluated next based on the knowledge gathered throughout previous evaluations. Accessing the quality of potential combinations will be done using the expected information (EI).Examples
population_table_training, peripheral_table = getml.datasets.make_numerical( random_state = 132) population_table_validation, _ = getml.datasets.make_numerical( random_state = 133) population_placeholder = population_table_training.to_placeholder() peripheral_placeholder = peripheral_table.to_placeholder() population_placeholder.join(peripheral_placeholder, join_key = "join_key", time_stamp = "time_stamp" ) feature_selector = getml.predictors.LinearRegression() predictor = getml.predictors.XGBoostRegressor() m = getml.models.MultirelModel( population = population_placeholder, peripheral = peripheral_placeholder, feature_selector = feature_selector, predictor = predictor, name = "multirel" ).send() param_space = { 'num_features': [80, 150], 'regularization': [0.3, 1.0], 'shrinkage': [0.1, 0.9] } g = getml.hyperopt.GaussianHyperparameterSearch( model = m, param_space = param_space, seed = int(datetime.datetime.now().timestamp()*100), session_name = 'test_search', n_iter = 45 ) g.fit( population_table_training = population_table_training, population_table_validation = population_table_validation, peripheral_tables = peripheral_table ) g.get_scores()
- Parameters
model (Union[
MultirelModel,RelboostModel]) – Base model used to derive all models fitted and scored during the hyperparameter optimization. Be careful in constructing it since only those parameters present in param_space too will be overwritten. It defines the data schema, any hyperparameters that are not optimized, and contains the predictor which will - depending on the parameter space - will be optimized as well.param_space (dict, optional) –
Dictionary containing numerical arrays of length two holding the lower and upper bounds of all parameters which will be altered in model during the hyperparameter optimization. To keep a specific parameter fixed, you have two options. Either ensure it is not present in param_space but in model, or set both the lower and upper bound to the same value. Note that all parameters in the
modelsandpredictorsdo have appropriate default values.If param_space is None, a default space will be chosen depending on the particular model and model.predictor. These default spaces will contain all parameters supported for the corresponding class and are listed below.
-
{ 'grid_factor': [1.0, 16.0], 'max_length': [1, 10], 'min_num_samples': [100, 500], 'num_features': [10, 500], 'regularization': [0.0, 0.01], 'share_aggregations': [0.01, 1.0], 'share_selected_features': [0.1, 1.0], 'shrinkage': [0.01, 0.4] }
-
{ 'max_depth': [1, 10], 'min_num_samples': [100, 500], 'num_features': [10, 500], 'reg_lambda': [0.0, 0.1], 'share_selected_features': [0.1, 1.0], 'shrinkage': [0.01, 0.4], }
LinearRegressionandLogisticRegression{ 'predictor_learning_rate': [0.5, 1.0], 'predictor_lambda': [0.0, 1.0] }
XGBoostClassifierandXGBoostRegressor{ 'predictor_n_estimators': [10, 500], 'predictor_learning_rate': [0.0, 1.0], 'predictor_max_depth': [3, 15], 'predictor_reg_lambda': [0.0, 10.0] }
To distinguish between the parameters belonging to the model from the ones associated with its predictor, the prefix ‘predictor_’ has to be added to the latter ones.
-
seed (Union[int,None], optional) – Seed used for the random number generator that underlies the sampling procedure to make the calculation reproducible. Due to nature of the underlying algorithm this is only the case if the fit is done without multithreading. To reflect this, a seed of None does represent an unreproducible and is only allowed to be set to an actual integer if both
num_threadsandn_jobsinstance variables of thepredictorandfeature_selectorin model - if they are instances of eitherXGBoostRegressororXGBoostClassifier- are set to 1. Internally, a seed of None will be mapped to 5543. Range: [0,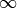 ]
]session_name (string, optional) –
Unique ID which will be both used as prefix for the
nameparameter of all models fitted during the hyperparameter optimization and directly inserted intosession_name. It will be used as a handle to load the constructed class from the getML engine.If session_name is empty, a default one based on the current date and time will be created.
Using a session_name all models trained in the engine during the hyperparameter optimization, which are based on the provided model, can be identified unambiguously.
n_iter (int, optional) –
Number of iterations in the hyperparameter optimization and thus the number of parameter combinations to draw and evaluate.
Literature on Gaussian processes suggests to have at least 10 different evaluations per dimension covered during the burn-in. Due to the characteristics of our feature engineering algorithms, we found that more relaxed lower limit of having n_iter - and not just the burn-in phase - be at least 10 times the number of dimensions in param_space still produces desirable results (while sticking to the rule of thumb of ratio_iter). Range: [4,
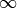 ]
]ratio_iter (float, optional) –
Percentage of the iterations used for the burn-in while the remainder will be used in training the Gaussian process. For a ratio_iter of 1.0 all iterations will be spend in the burn-in period resulting in an equivalence of this class to
LatinHypercubeSearchorRandomSearch- depending on surrogate_burn_in_algorithm. Range: [0, 1]As a rule of thumb at least 70 percent of the evaluation should be spent during the burn-in. The more comprehensive the exploration of the param_space during the burn-in, the less likely the Gaussian process to get stuck in bad local minima. While the training of the process itself uses a trade-off between exploration and exploitation and is thus able to escape these minima itself, it could very well stick in there for a dozen iterations.
optimization_algorithm (string, optional) –
Determines the optimization algorithm used for the local search in the optimization of the expected information (EI).
Right now two choices are supported: ‘nelderMead’, a gradient-free downhill simplex method, and ‘bfgs’, a quasi-Newton method relying on both the negative log-likelihood and its gradient. We found the ‘nelderMead’ algorithm to work slightly more reliable.
optimization_burn_in_algorithm (string, optional) –
Specifies the algorithm used to draw initial points in the burn-in period of the optimization of the expected information (EI).
For a detailed explanation of the two possible choices - ‘latinHypercube’ and ‘random’ - please have a look in the documentation of the
LatinHypercubeSearchandRandomSearchclass. In general, the ‘latinHypercube’ is recommended since it more likely to resulting in a good coverage of the parameter space.optimization_burn_ins (int, optional) –
Number of random evaluation points used during the burn-in of the minimization of the expected information (EI).
After the surrogate model - the Gaussian process - was successfully fitted to the previous parameter combination, one is able to calculate the EI for a given point. In order to now get to the next combination, the EI has to be maximized over the whole parameter space. But this problem suffers heavily from local minima too. So, we have to start the optimization leading to the suggestion of the next parameter combination with evaluating the EI at various, random points - using optimization_burn_in_algorithm - and to use a local search - specified in optimization_algorithm - on the best result. Range: [3,
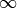 ]
]surrogate_burn_in_algorithm (string, optional) –
Specifies the algorithm used to draw new parameter combinations during the burn-in period.
For a detailed explanation of the two possible choices - ‘latinHypercube’ and ‘random’ - please have a look in the documentation of the
LatinHypercubeSearchandRandomSearchclass. In general, the ‘latinHypercube’ is recommended since it more likely to resulting in a good coverage of the parameter space.gaussian_kernel (string, optional) –
Specifies the 1-dimensional kernel of the Gaussian process which will be used along each dimension of the parameter space. All of the choices below will result in continuous sample paths and their main difference is the degree of smoothness of the results with ‘exp’ yielding the least and ‘gauss’ yielding the most smooth ones.
’exp’
A Exponential kernel yielding non-differentiable sample paths.
’matern32’
A Matérn 3/2 kernel yielding once-differentiable sample paths.
’matern52’
A Matérn 5/2 kernel yielding twice-differentiable sample paths.
’gauss’
A Gaussian kernel yielding analytic (infinitely–differentiable) sample paths.
gaussian_optimization_algorithm (string, optional) –
Determines the optimization algorithm used for the local search in the fitting of the Gaussian process to the previous parameter combinations.
Right now two choices are supported: ‘nelderMead’, a gradient-free downhill simplex method, and ‘bfgs’, a quasi-Newton method relying on both the negative log-likelihood and its gradient. We found the ‘nelderMead’ algorithm to work slightly more reliable.
gaussian_optimization_burn_in_algorithm (string, optional) –
Specifies the algorithm used to draw new parameter combinations during the burn-in period of the optimization of the Gaussian process.
For a detailed explanation of the two possible choices - ‘latinHypercube’ and ‘random’ - please have a look in the documentation of the
LatinHypercubeSearchandRandomSearchclass. In general, the ‘latinHypercube’ is recommended since it more likely to resulting in a good coverage of the parameter space.gaussian_optimization_burn_ins (int, optional) – Number of random evaluation points used during the burn-in of the fitting of the Gaussian process. Range: [3,
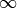 ]
]
- Raises
KeyError – If an unsupported instance variable is encountered (via
validate()).TypeError – If any instance variable is of wrong type (via
validate()).ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical) (via
validate()).ValueError – If not
predictoris present in the provided model.
Note
What’s the incentive behind using a Bayesian hyperparameter optimization anyway and how does it work?
Our overall goal is to get the best hyperparameter combination in order to perform the best prediction possible. To rephrase it in mathematical terms, we want to minimize the negative log-likelihood of an objective function representing the performance of the feature engineering algorithm, the feature selector, and predictor (measured using a particular score) given a set of data. But the surface of this negative log-likelihood is not convex and contains many local minima. We, thus, need to use a global optimization scheme. First of all we sample random points in the parameter space, evaluate the objective functions at all those sites, and, finally, start a well-known and tested local optimization routine, e.g. Nelder-Mead, at the best-performing combination. The initial point in our parameter space used to start the optimization from will be the parameters of the provided model - either the ones you chose manually or the default ones in the
MultirelModelorRelboostModelconstructors, which we will call the base model from here on.But on top of having local minima, the objective function has a far worse property: it is very expensive to evaluate. Local optimization algorithms can easily require over one hundred iterations to converge which usually is not an issue (e.g. minimizing the negative log-likelihood of a distribution function on, let’s say, 1000 data points only takes about 10ms on modern computers). But if evaluating the objective function involves performing a multi-stage fit of various machine learning algorithms to a large amount of data, each iteration can take minutes or even longer. In such a scenario even the simple task of performing a local minimization very quickly becomes computationally infeasible.
This is where Bayesian hyperparameter optimization enters the stage. Its idea is to not fit the negative log-likelihood of the objective function directly but, instead, to approximate it with a surrogate model - the Gaussian process [Rasmussen06] - and to fit the approximation instead. By doing so we trade evaluation time - since the surrogate is much more cheap to evaluate - in for accuracy - since we are only dealing with an approximation of the real objective function.
The first part of our global optimization scheme (sampling the parameter space), becomes a lot more crucial since not just the quality of the starting points for the local optimization but, even more important, the approximation by the Gaussian process does intimately depend on the number and distribution of previous evaluations. Without a good coverage of the parameter space the Gaussian process will not resemble its target properly and the results of the local optimization will be poorly.
Fortunately, we can do better than simply drawing random parameter combinations, fitting a single Gaussian process afterwards, and returning its minimum. The second core idea of the Bayesian hyperparameter optimization is to redesign the global optimization for better and more efficient performance. The local optimization will be replaced by an iterative scheme in which the surrogate is fitted to all previous parameter combinations and used to find the most promising combination to evaluate next. As a measure of quality for the next point to evaluate - also called acquisition function in the Bayesian optimization community - we use the expected information (EI) [Villemonteix09]. It measures how much improvement with respect to the current minimum of the negative log-likelihood function is expected when evaluating a particular additional point given all previous evaluations. An immense benefit of using the maximum of the EI (or other acquisition functions) calculated for the Gaussian process over the raw minimum of the surrogate is that they provide a trade-off between exploration and exploitation of the parameter space and are thus able to efficiently fit the objective function “on their own”. The EI itself we also have to optimize using a global scheme throughout the whole parameter space. But since this is done on top of the Gaussian process, it is quite fast.
To summarize, the optimization starts by drawing a number of points within the parameter space at random and using them to fit and score the model. Next, a Gaussian process is fitted to all parameter combinations/calculated score pairs. Using it the most likely position of the optimal parameter combination can be determined by optimizing the EI. The model corresponding to this combination is fitted and scored by the getML engine and, again, a Gaussian process is fitted to all evaluations. This procedure will be continued until the maximum number of iterations n_iter is reached. As a result you get a list of all fitted variants of the base model as well as their calculated scores. Note, however, that the final parameter combination calculated based on the models in the provided list will not be returned. Since the EI is a trade-off between exploration and exploitation, the last combination does not have to be the optimal one and we only keep those models we know the performance (score) of.
The algorithm occasionally does evaluate quite a number of evaluations (sometimes several dozen) in what appears to be the global minimum while in reality it got stuck in a local one. The possibility for such an event to happen is particularly high for high-dimensional spaces or/and too short burn-in periods. In time the algorithm will be able to escape the local minima and approach the global one. But instead of increasing the number of surrogate evaluation, we recommend to perform a more thorough burn-in period instead.
References
Carl Edward Rasmussen and Christopher K. I. Williams, MIT Press, 2006
Julien Villemonteix, Emmanuel Vazquez, and Eric Walter, 2009
Methods Summary
fit(population_table_training, …[, score])Launches the hyperparameter optimization.
Get a list of all models fitted during the hyperparameter optimization.
Get a dictionary of the score corresponding to all models fitted during the hyperparamer optimization.
validate()Checks both the types and the values of all instance variables and raises an exception if something is off.
Methods Documentation
-
fit(population_table_training, population_table_validation, peripheral_tables, score=None)¶ Launches the hyperparameter optimization.
The optimization itself will be done by the getML software and this function returns immediately after constructing the request and checking whether population_table_training and population_table_validation do hold the same column names using
_validate_colnames().In every iteration of the hyperparameter optimization a new set of hyperparameters will be drawn from the param_space member of the class, those particular parameters will be overwritten in the base model and it will be renamed, fitted, and scored. How the hyperparameters themselves are drawn depends on the particular class of hyperparameter optimization.
The provided
DataFramepopulation_table_training, population_table_validation and peripheral_tables must be consistent with thePlaceholdersprovided when constructing the base model.- Parameters
population_table_training (
DataFrame) – The population table that models will be trained on.population_table_validation (
DataFrame) – The population table that models will be evaluated on.peripheral_tables (
DataFrame) – The peripheral tables used to provide additional information for the population tables.score (string, optional) –
The score with respect to whom the hyperparameters are going to be optimized.
Possible values for a regression problem are:
Possible values for a classification problem are:
cross_entropy(default)
- Raises
TypeError – If any of population_table_training, population_table_validation or peripheral_tables is not of type
DataFrame.KeyError – If an unsupported instance variable is encountered (via
validate()).TypeError – If any instance variable is of wrong type (via
validate()).ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical) (via
validate()).
-
get_models()¶ Get a list of all models fitted during the hyperparameter optimization.
- Returns
List of all models fitted during the hyperparameter optimization.
- Return type
list
- Raises
Exception – If the engine yet reports back that the operation was not successful.
KeyError – If an unsupported instance variable is encountered (via
validate()).TypeError – If any instance variable is of wrong type (via
validate()).ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical) (via
validate()).
-
get_scores()¶ Get a dictionary of the score corresponding to all models fitted during the hyperparamer optimization.
- Returns
All score fitted during the hyperparameter optimization. Each field adheres the following scheme:
{"model-name": {"accuracy": [list_of_scores], "auc": [list_of_scores], "cross_entropy": [list_of_scores], "mae": [list_of_scores], "rmse": [list_of_scores], "rsquared": [list_of_scores]}
For more information regarding the scores check out
getml.models.scores(listed under ‘Variables’).- Return type
dict
- Raises
Exception – If the engine yet reports back that the operation was not successful.
KeyError – If an unsupported instance variable is encountered (via
validate()).TypeError – If any instance variable is of wrong type (via
validate()).ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical) (via
validate()).
-
validate()¶ Checks both the types and the values of all instance variables and raises an exception if something is off.
Examples
population_table, peripheral_table = getml.datasets.make_numerical() population_placeholder = population_table.to_placeholder() peripheral_placeholder = peripheral_table.to_placeholder() population_placeholder.join(peripheral_placeholder, join_key = "join_key", time_stamp = "time_stamp" ) feature_selector = getml.predictors.LinearRegression() predictor = getml.predictors.XGBoostRegressor() m = getml.models.MultirelModel( population = population_placeholder, peripheral = peripheral_placeholder, feature_selector = feature_selector, predictor = predictor, name = "multirel" ).send() param_space = { 'num_features': [80, 150], 'regularization': [0.3, 1.0], 'shrinkage': [0.1, 0.9] } g = getml.hyperopt.GaussianHyperparameterSearch( model = m, param_space = param_space, seed = int(datetime.datetime.now().timestamp()*100), session_name = 'test_search' ) g.optimization_burn_ins = 240 g.model.num_threads = 2 g.validate()
- Raises
KeyError – If an unsupported instance variable is encountered.
TypeError – If any instance variable is of wrong type.
ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical).
Note
This method is called at end of the __init__ constructor and every time a method is communicating with the getML engine.
To directly access the validity of single or multiple parameters instead of the whole class, you can used
getml.helpers.validation.validate_hyperopt_parameters().