MultirelModel¶
-
class
getml.feature_learning.MultirelModel(aggregation=None, allow_sets=True, delta_t=0.0, grid_factor=1.0, loss_function='SquareLoss', max_length=4, min_num_samples=1, num_features=100, num_subfeatures=5, num_threads=0, regularization=0.0, round_robin=False, sampling_factor=1.0, seed=None, share_aggregations=0.25, share_conditions=1.0, shrinkage=0.0, silent=True, use_timestamps=True)¶ Bases:
getml.feature_learning.feature_learner._FeatureLearnerFeature learning based on Multi-Relational Decision Tree Learning.
MultirelModelautomates feature learning for relational data and time series. It is based on an efficient variation of the Multi-Relational Decision Tree Learning (MRDTL) algorithm and uses the getML Multirel algorithm.For more information on the underlying feature learning algorithm, check out the User guide.
- Parameters
aggregation (List[
aggregations], optional) –Mathematical operations used by the automated feature learning algorithm to create new features.
Possible options:
AvgCountCountDistinctCountMinusCountDistinctMaxMedianMinStddevSumVar
allow_sets (bool, optional) –
Multirel can summarize different categories into sets for producing conditions. When expressed as SQL statements these sets might look like this:
t2.category IN ( 'value_1', 'value_2', ... )
This can be very powerful, but it can also produce features that are hard to read and might be prone to overfitting when the sampling_factor is too low.
delta_t (float, optional) –
Frequency with which lag variables will be explored in a time series setting. When set to 0.0, there will be no lag variables.
For more information please refer to Time series. Range: [0,
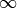 ]
]grid_factor (float, optional) – Multirel will try a grid of critical values for your numerical features. A higher grid_factor will lead to a larger number of critical values being considered. This can increase the training time, but also lead to more accurate features. Range: (0,
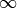 ]
]loss_function (
loss_functions, optional) – Objective function used by the feature learning algorithm to optimize your features. For regression problems useSquareLossand for classification problems useCrossEntropyLoss.max_length (int, optional) –
The maximum length a subcondition might have. Multirel will create conditions in the form
(condition 1.1 AND condition 1.2 AND condition 1.3 ) OR ( condition 2.1 AND condition 2.2 AND condition 2.3 ) ...
Using this parameter you can set the maximum number of conditions allowed in the brackets. Range: [0,
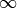 ]
]min_num_samples (int, optional) – Determines the minimum number of samples a subcondition should apply to in order for it to be considered. Higher values lead to less complex statements and less danger of overfitting. Range: [1,
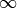 ]
]num_features (int, optional) – Number of features generated by the feature learning algorithm. Range: [1,
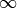 ]
]num_subfeatures (int, optional) – The number of subfeatures you would like to extract in a subensemble (for snowflake data model only). See The snowflake schema for more information. Range: [1,
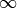 ]
]num_threads (int, optional) – Number of threads used by the feature learning algorithm. If set to zero or a negative value, the number of threads will be determined automatically by the getML engine. Range: [
 ,
, 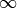 ]
]regularization (float, optional) – Most important regularization parameter for the quality of the features produced by Multirel. Higher values will lead to less complex features and less danger of overfitting. A regularization of 1.0 is very strong and allows no conditions. Range: [0, 1]
round_robin (bool, optional) – If True, the Multirel picks a different aggregation every time a new feature is generated.
sampling_factor (float, optional) – Multirel uses a bootstrapping procedure (sampling with replacement) to train each of the features. The sampling factor is proportional to the share of the samples randomly drawn from the population table every time Multirel generates a new feature. A lower sampling factor (but still greater than 0.0), will lead to less danger of overfitting, less complex statements and faster training. When set to 1.0, roughly 2,000 samples are drawn from the population table. If the population table contains less than 2,000 samples, it will use standard bagging. When set to 0.0, there will be no sampling at all. Range: [0,
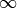 ]
]seed (Union[int,None], optional) – Seed used for the random number generator that underlies the sampling procedure to make the calculation reproducible. Due to nature of the underlying algorithm this is only the case if the fit is done without multithreading. To reflect this, a seed of None does represent an unreproducible and is only allowed to be set to an actual integer if both num_threads and
n_jobsinstance variables of the predictor and feature_selector - if they are instances of eitherXGBoostRegressororXGBoostClassifier- are set to 1. Internally, a seed of None will be mapped to 5543. Range: [0,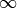 ]
]share_aggregations (float, optional) – Every time a new feature is generated, the aggregation will be taken from a random subsample of possible aggregations and values to be aggregated. This parameter determines the size of that subsample. Only relevant when round_robin is False. Range: (0, 1]
share_conditions (float, optional) – Every time a new column is tested for applying conditions, it might be skipped at random. This parameter determines the probability that a column will not be skipped. Range: [0, 1]
shrinkage (float, optional) – Since Multirel works using a gradient-boosting-like algorithm, shrinkage (or learning rate) scales down the weights and thus the impact of each new tree. This gives more room for future ones to improve the overall performance of the model in this greedy algorithm. Higher values will lead to more danger of overfitting. Range: [0, 1]
silent (bool, optional) – Controls the logging during training.
use_timestamps (bool, optional) –
Whether you want to ignore all elements in the peripheral tables where the time stamp is greater than the one in the corresponding elements of the population table. In other words, this determines whether you want add the condition
t2.time_stamp <= t1.time_stamp
at the very end of each feature. It is strongly recommend to enable this behavior.
- Raises
TypeError – If any of the input arguments is of wrong type.
KeyError – If an unsupported instance variable is encountered.
TypeError – If any instance variable is of wrong type.
ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical).
Example
population_placeholder = getml.data.Placeholder("population") order_placeholder = getml.data.Placeholder("order") trans_placeholder = getml.data.Placeholder("trans") population_placeholder.join(order_placeholder, join_key="account_id") population_placeholder.join(trans_placeholder, join_key="account_id", time_stamp="date") feature_selector = getml.predictors.XGBoostClassifier( reg_lambda=500 ) predictor = getml.predictors.XGBoostClassifier( reg_lambda=500 ) agg = getml.feature_learning.aggregations feature_learner = getml.feature_learning.MultirelModel( aggregation=[ agg.Avg, agg.Count, agg.Max, agg.Median, agg.Min, agg.Sum, agg.Var ], num_features=60, loss_function=getml.feature_learning.loss_functions.CrossEntropyLoss ) pipe = getml.pipeline.Pipeline( tags=["multirel", "31 features"], population=population_placeholder, peripheral=[order_placeholder, trans_placeholder], feature_learners=feature_learner, feature_selectors=feature_selector, predictors=predictor, share_selected_features=0.5 ) pipe.check( population_table=population_train, peripheral_tables={"order": order, "trans": trans} ) pipe = pipe.fit( population_table=population_train, peripheral_tables={"order": order, "trans": trans} ) in_sample = pipe.score( population_table=population_train, peripheral_tables={"order": order, "trans": trans} ) out_of_sample = pipe.score( population_table=population_test, peripheral_tables={"order": order, "trans": trans} )
Methods Summary
validate([params])Checks both the types and the values of all instance variables and raises an exception if something is off.
Methods Documentation
-
validate(params=None)¶ Checks both the types and the values of all instance variables and raises an exception if something is off.
- Parameters
params (dict, optional) – A dictionary containing the parameters to validate. If not is passed, the own parameters will be validated.
- Raises
KeyError – If an unsupported instance variable is encountered.
TypeError – If any instance variable is of wrong type.
ValueError – If any instance variable does not match its possible choices (string) or is out of the expected bounds (numerical).